One of the most used methods for clustering data is the K-means method. This is an unsupervised method for categorizing data into various cluster groups. The number of groups in the data is represented by K. K-Means, a Hard and Flat Clustering Method. As we mentioned at the start of this section, hard clustering refers to the fact that not every data point in the clusters presently makes each cluster unique. These clusters don’t overlap. K-Means, a Flat clustering technique, isn’t hierarchical. Each cluster is not a member of any other cluster.
How The Algorithm Work
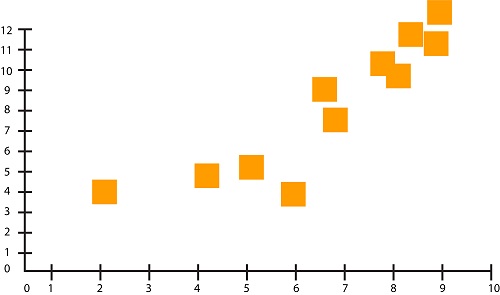
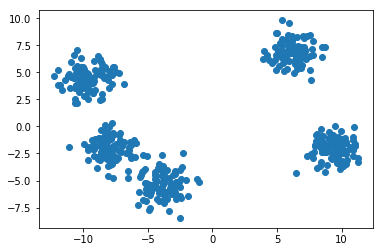
Let us say that we have two features in our dataset. This figure can be seen when plotted on graphs:
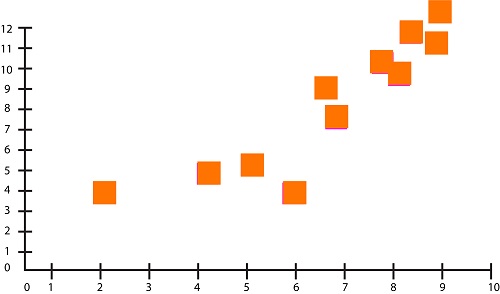
We will now take an arbitrary value of k. To create two groups from the above data, the algorithm picks two random points to serve as the centroid. It then calculates Euclidean distances to the centroid from all other data points. Let’s suppose that the centroid to the right has the label “1”, while the one to the left has “2”.
After measuring the distances between all the data points and the centroid, the algorithm associates every data point with a specific centroid based on its proximity.
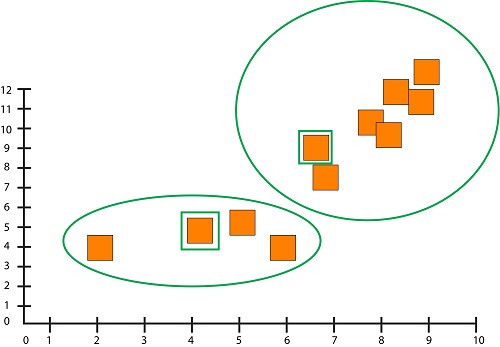
K-means algorithm works iteratively. It updates the centroids by taking the average of all data points assigned to each centroid’s group.
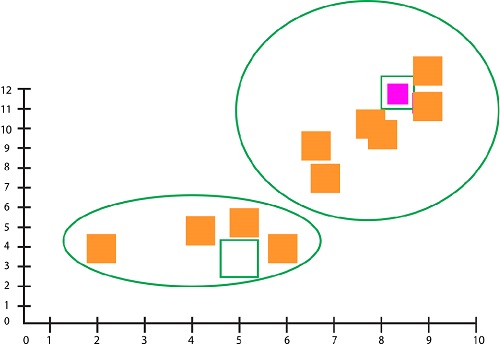
It is now in a new position. It continues the process as before, but this time it computes the mean and updates the position of the centroid. After updating the centroid, no data points change the clusters. This is the point at convergence.
Therefore, K-means is very different from other supervised learning algorithms. We had labels and updated the algorithm based on the error we received in the output.
Mathematical Calculation
You can understand the above algorithm by doing some calculations. We can also understand how Euclidean distances are calculated and how centroids are updated in order to form clusters.
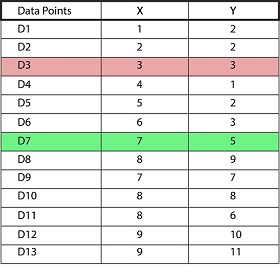
We would like to form two clusters (k=2) using a dataset that has two features (X and Y) with 13 data points.
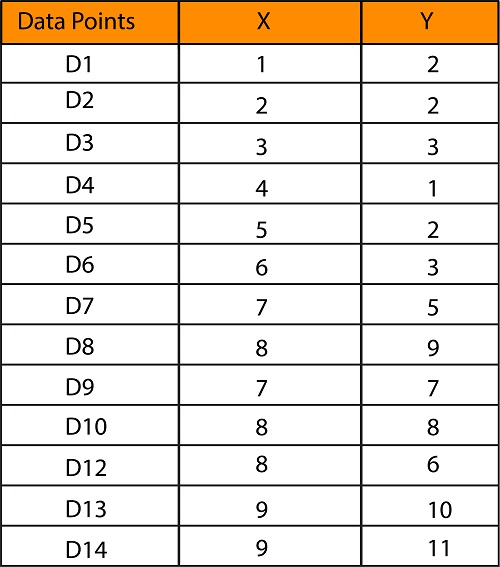
If plotted using x-y coordinates, you get the scatter plot we saw earlier.
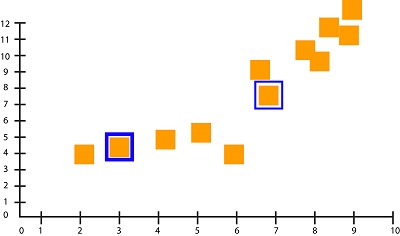
Select the random data points to be the centroid. For example, we choose Datapoint 3 as Centroid 1 (Cluster Liable = 1) and Datapoint 7 as Centroid 2(Cluster Label = 2). These two data points serve as our initial random centroids.
You can also mark them on your graph to get a better view of where they are.
Next, calculate the distance between these two centroids and all of the data points. Here we will use the most commonly used distance metric, the Euclidean distance.
We have the following table after calculating the Euclidean distance to the centroid from each data point:
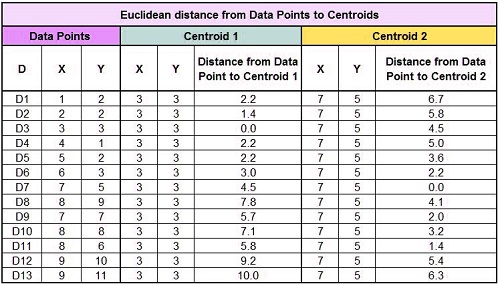
Now, we must compare the Euclidean distances between each data point (D1) and the two centroids. We can see, for example, that D1 is closer to Centroid 1 (6.7) than D1. We can therefore assign this data point (Centroid 1) to Cluster Label = 1.
We do the same thing for all the samples, assigning them to a cluster based on their proximity to the centroid.
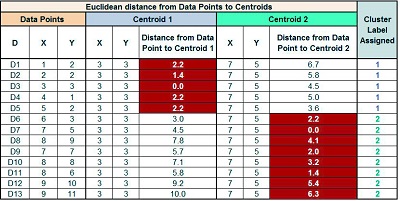
If we make a cluster in the graph, you will notice that the data at the graph’s left-bottom form a Cluster (Cluster Name 1), and the data at the top of the graph form a Cluster (Cluster Name 2)
As we’ve already mentioned, updating the centroids requires we take the mean of all data points associated with each centroid’s Cluster. We take the average of all data points in Cluster 1 to find the updated Centroid 1. For Centroid 2, we use the same procedure.
We then calculate the distance from each updated centroid to all data points. Finally, we compare these distances to assign clusters. Cluster Label 1 was previously assigned to D6. Now, it is designated as Cluster Label 2!
As we mentioned, the algorithm repeats the above process until it reaches a point of convergence. That is until cluster labels are no longer resigned to updating the centroids. The following calculations can be done if we update the centroids:
We discover that the new centroids are Centroid 1 (3,2.22) and Centroid 2 (8.8).
If we compare the distances, we will find that the convergence point has been reached. No cluster labels are being updated, so we can stop the process from arriving at the final clusters.
Determining how many K
The most important parameter is the value k. There are many ways to find K.
Profiling Approach
This method allows us to identify the characteristics of each segment. It is possible if we have good domain knowledge and are able to spend a lot of time. These are multiple values for k. After analyzing each cluster, the segment with the best meaning is selected.
Suppose we have a data set and run the algorithm with k=3,4, 5, 6, 6, and 7, respectively. We get the following results.
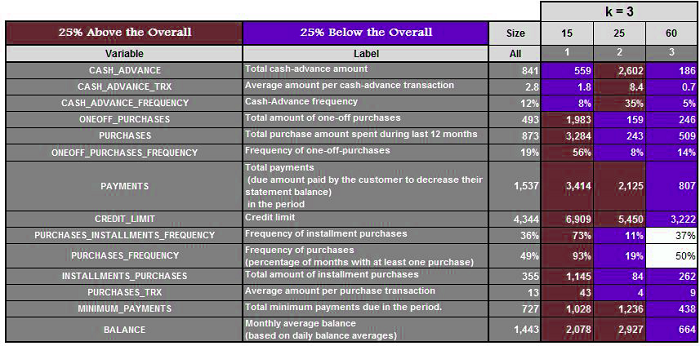
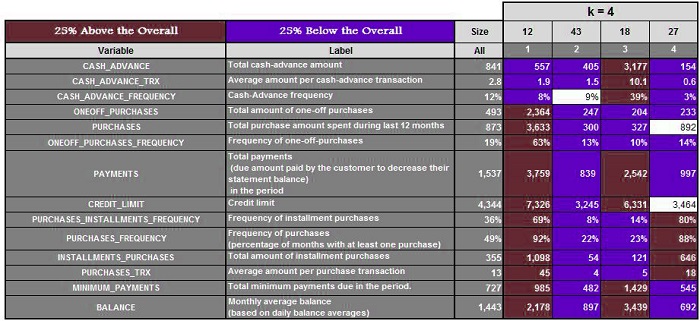
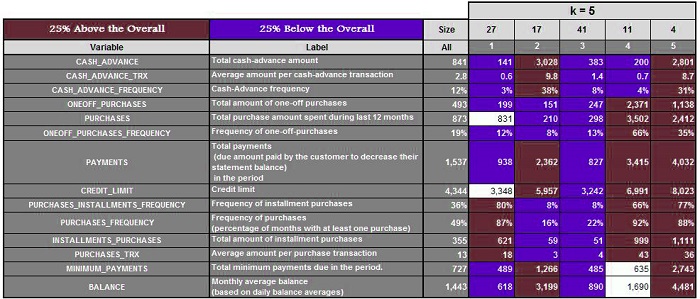
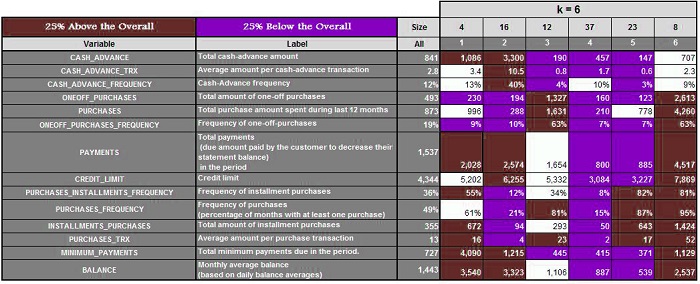
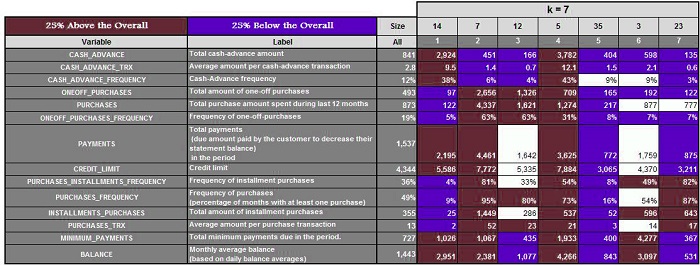
Three clusters are created when k=3. Next, we calculate the mean of these features and highlight the ones that are 25% above and 25% below the average. For K=4,5,6, and 7, we do the same.
It is now possible to analyze each cluster for every value of K and determine which value provides us with the most meaningful clusters. (In our case, it was k=5) This method is time-consuming and can fail if you don’t have any domain knowledge or don’t know the range where k should be.
Elbow Analysis
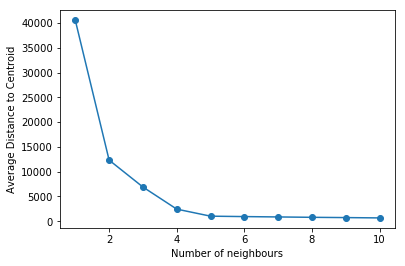
The Elbow Analysis is another popular method. Using this method, we calculate the average distance between the data points and their centroid. As the number of centroids increases, so does the distance between the data points and the centroid. Multiple values of k can be used. As shown below, plot the graphs. Find the slope decreasing and the average distance leveling out for the value of K.
The Silhouette Coefficient, which considers the largest coefficient and the value of K, is another way to validate the value. You can also use other metrics like ANOVA, SC Test, or Dendrograms.
Role in Seed
The algorithm’s goal is not to reduce errors (as we are working in an unsupervised environment) but to minimize the total distance from each centroid to all data points. This algorithm is concerned with intercluster distance. We don’t care much about distances between clusters. The problem with this clustering approach is that we select the centroids randomly. No matter what random centroids we use, we will all converge. However, we converge to a locally optimal, which means we can have different results if we start from different centroids. In the context of coding, this is the way we start the random selection process. This is done by giving a random seed. When we change the seed, the value of k from an elbow analysis can differ. This indicates that the initial random centroids influence the convergence. Manually, you can run the k = clustering for n number k times for m numbers of times each time with different random seeds. After that, average the results of your elbow analysis to get the right number.
Preprocessing is Required for K-means Clustering
Outlier Treatment
Outlier Therapy, like all distance-based techniques, must be performed on data before it is used for k–means clustering. You can read about the various outlier treatment options in the blog: Outlier Treat and Anomaly detection.
Missing value
The missing values treatment blog has many ways to handle missing values.
Rescaling Information
Feature Scaling mentioned that the distance-based algorithm should be rescaled to make sense of the algorithm.
Enoding
Additionally, features must be numerical. One hot-encoding/dummy variable generation will be necessary if data contains categorical variables. This was discussed in the blog.
Dimensionality Reduction
There are many Dimensionality Reduce techniques, such as a PCA, that can reduce the number of features that are not needed and can make K-means’ output less meaningful.
